- Tue 17 September 2013
- notebook
- Jason K. Moore
- #notebook, #filtering, #zotero, #plone
Today's task list:
- [x] Review Tarun's PRs
- [x] Work on the website content and structure
- [x] Do CITI course
- [x] Lab meeting
- [x] investigate displaying publications on the website
- [] Work on BMD papers
- [] Work on parsing the walking data
- [] Working on wrapping Ton's walking models
- [] Book hotel for BMD
- [] Post update about BMD copyright
- [] Finish reading the van der Kooij paper
- [] See if our controller can drive an OpenSim model or Ton's 2D model
- [] Wrap the HBM C code
- [] Duplicate website backups on a S3 bucket
- [] Work on the website theme
- [] Make generic settings on the lab website
- [] Setup workflow for website for public documents
- [] Investigate ways to display publication lists on the website
- [] Embed the google calendar on the website
- [] Review the TODO items on the Yeadon paper
- [] Do FERPA course, due Sept 20
- [] Write up database proposal
- [] Try out CSympy with some mechanics problems
- [] Email Mounir about teaching
Butterworth Filters
Sandy noticed that Luuk may not have been setting the crossover frequency correctly in his report. In the Matlab style digital Butterworth filter design you typically set the wn argument to:
desired filter cutoff frequency nyquist frequency of the signal
This is a normalized cutoff frequency. Our system samples at 100 hz max due to the motion capture data limits. You can pull in data at higher sample rates but then you get repeated values for the mocap signals.
I wrote this little script to show what happens if you select a nyquist frequency other than the actual sample rate of the signal you want to filter:
https://gist.github.com/moorepants/6597067
The resulting plot is:
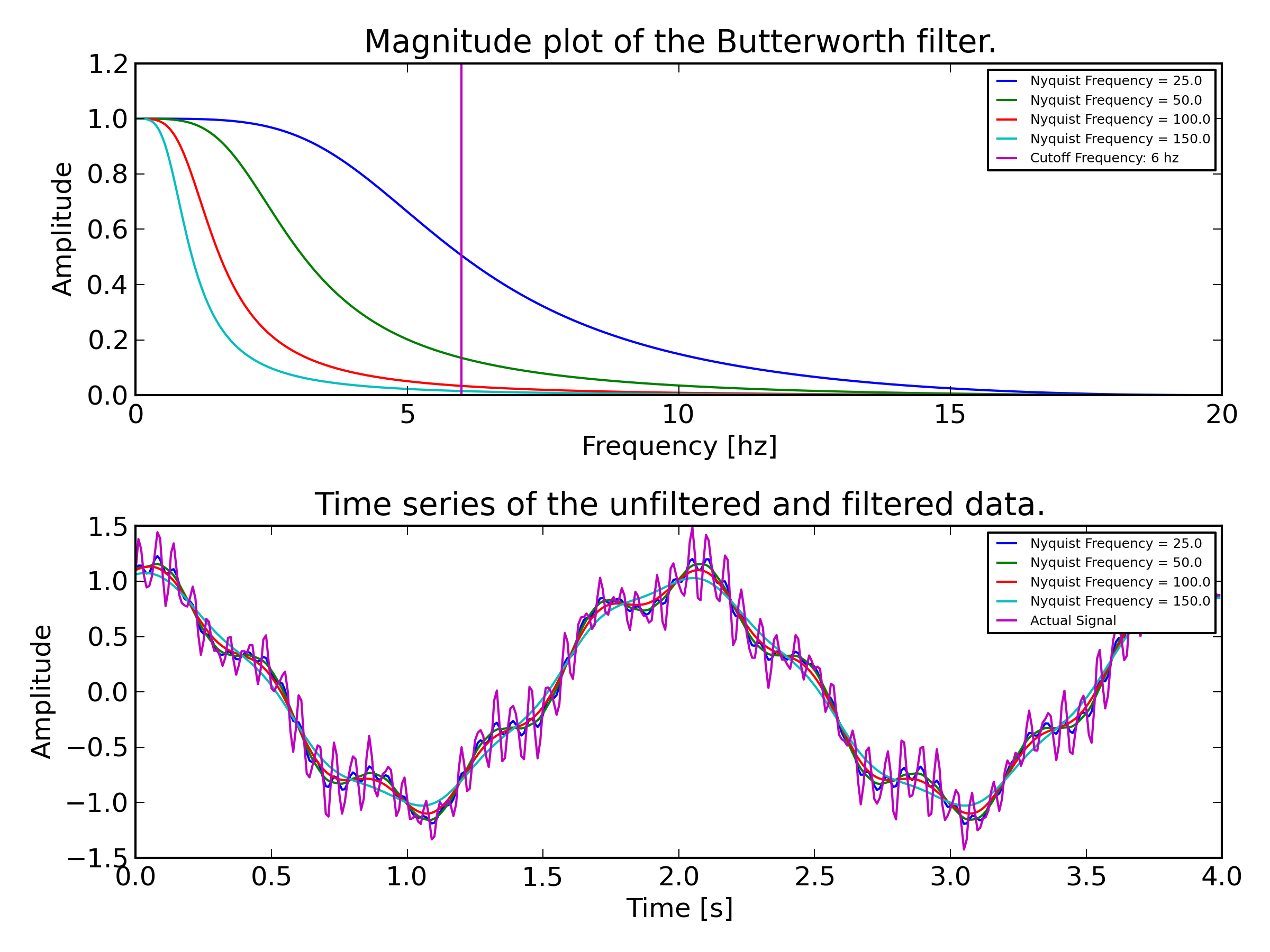
If you choose a larger Nyquist frequency than the signal, then you push the passband frequency to a lower value. Luuk may have done this incorrectly in his report.
CITI Test
Woohoo, finished the IRB and ethical research tests. They were long but informative. Some seemed really outdated. I posted some details to my G+ account about passages that I thought weren't true.
Plone Bibliography Support
Three options that I'm looking into:
- Install Products.CMFBibliographyAT and users can create bibliography folders and manually add items.
- Create a custom content type that pulls in HTML from a public Zotero group and displays it.
- Create a button on the TinyMCE editor which, when clicked, prompts the user for a public zotero group and collection id, and embeds the content from the resulting zotero api call into the page.
I could setup some super simple pages that display stuff served up by pyzotero on our server outside of plone, and them iframe it into a plone page. But that wouldn't have styling and would probably suck.
Zotero gives atom and xhtml standard api calls:
http://www.zotero.org/support/dev/server_api/v2/read_requests#example_requests_and_responses
The atom feed can be read in any feed reader (i.e. a Plone portlet).
If I need to do the zotero api calls from Python in Plone there is this lib:
https://github.com/urschrei/pyzotero
Here is an API call to a public Zotero group I set up:
https://api.zotero.org/groups/212385/collections/JWGBWM9P/items?format=bib
It sends back some simple xhtml.
How to add plugins to TinyMCE:
http://www.tinymce.com/wiki.php/TinyMCE3x:Creating_a_plugin
Plone products that have TinyMce plugins:
https://pypi.python.org/pypi?%3Aaction=search&term=collective.tinymce&submit=search
Very simple view of zotero data in Plone:
https://github.com/collective/collective.pece/blob/master/collective/pece/browser/zotero_view.py
